Difference between revisions of "Spectra-PKA"
m |
Mark Gilbert (Talk | contribs) |
||
| Line 1: | Line 1: | ||
<tt>SPECTRA-PKA</tt> is a command-line driven programme for calculating the expected primary knock-on atom (PKA) spectra for a given target nuclide under neutron or charged particle irradiation. NJOY-processed recoil matrices must be provided as the input nuclear data for each nuclide and reaction channel of interest. <tt>SPECTRA-PKA</tt> will read the nuclear data and collapse the data for each reaction channel in a file with a user-defined energy spectrum of incident particles. <tt>SPECTRA-PKA</tt> outputs the resulting PKA spectra for each reaction channel read-in, as well as summed PKA distributions for each different recoiling nuclide or element, including both the typically heavy residuals and the secondary-emitted light gas particles. Crucially, <tt>SPECTRA-PKA</tt> has the ability to process data, in a single run, for a complex material containing many different target species. The user must specify the atomic percentage for each target and the appropriate recoil cross section file, but then <tt>SPECTRA-PKA</tt> will automatically read-in and process each file and create global average (including as a function of isotope and element) PKA distributions. This feature, in particular, is a significant advancement over what was possible before (such as with <tt>SPECTER</tt> [1985] from ANL), where typically PKA distributions were only provided for single elements (and not separated by reaction channel) and based on only one nuclear data library (ENDF/B-V in the case of <tt>SPECTER</tt>). It allows, for example, the user to investigate the variation in PKA distributions as a function of time under irradiation, where a materials composition may change due to transmutation. | <tt>SPECTRA-PKA</tt> is a command-line driven programme for calculating the expected primary knock-on atom (PKA) spectra for a given target nuclide under neutron or charged particle irradiation. NJOY-processed recoil matrices must be provided as the input nuclear data for each nuclide and reaction channel of interest. <tt>SPECTRA-PKA</tt> will read the nuclear data and collapse the data for each reaction channel in a file with a user-defined energy spectrum of incident particles. <tt>SPECTRA-PKA</tt> outputs the resulting PKA spectra for each reaction channel read-in, as well as summed PKA distributions for each different recoiling nuclide or element, including both the typically heavy residuals and the secondary-emitted light gas particles. Crucially, <tt>SPECTRA-PKA</tt> has the ability to process data, in a single run, for a complex material containing many different target species. The user must specify the atomic percentage for each target and the appropriate recoil cross section file, but then <tt>SPECTRA-PKA</tt> will automatically read-in and process each file and create global average (including as a function of isotope and element) PKA distributions. This feature, in particular, is a significant advancement over what was possible before (such as with <tt>SPECTER</tt> [1985] from ANL), where typically PKA distributions were only provided for single elements (and not separated by reaction channel) and based on only one nuclear data library (ENDF/B-V in the case of <tt>SPECTER</tt>). It allows, for example, the user to investigate the variation in PKA distributions as a function of time under irradiation, where a materials composition may change due to transmutation. | ||
| − | |||
| − | <tt>SPECTRA-PKA</tt> is executed from the command-line. By default, the programme expects to find an input file called | + | == Installation == |
| + | <tt>SPECTRA-PKA</tt> comes with a simple makefile, requiring GNU make and a compatible Fortran compiler. It has been tested with multiple operating systems and compiler permutations, including the standard GNU Fortran compiler. | ||
| + | |||
| + | == Usage == | ||
| + | <tt>SPECTRA-PKA</tt> is executed from the command-line. By default, the programme expects to find an input file called specter.input in the execution folder, but this behaviour can be over-ridden by specifying the name of an input file on the command-line. The input file provides the user with the option to over-ride the default values of the various code words that <tt>SPECTRA-PKA</tt> relies upon to control its execution. The input file should be a text file but is largely free from other formatting requirements. Code word values are specified in any order, one per line, via syntax of type: | ||
<pre><code word name> = <code word value></pre> | <pre><code word name> = <code word value></pre> | ||
| − | |||
| − | |||
== Code words == | == Code words == | ||
| + | A complete list of code words (with explanation) can be found on the pdf readme on the git respository for <tt>SPECTRA-PKA</tt>. An alternative syntax allowing an easier description of a complex alloy (containing multiple target species) is also described. | ||
| − | + | == Input PKA data download == | |
| − | + | <tt>SPECTRA-PKA</tt> uses NJOY-generated recoil cross section matrices. The output files required from NJOY are non-standard and are produced by a slightly modified version of the GROUPR module within NJOY. An evolving selection of databases produced using various international reaction cross section libraries (for various incident particle types) have been pre-calculated and are available to download as compressed tar archives from [http://www.ccfe.ac.uk/FISPACT-II/nuclear_data/PKA here] (links are also provided in a detailed listing of nuclear data available for FISPACT-II at [https://fispact.ukaea.uk/nuclear-data/downloads nuclear data downloads]). These can be used immediately with <tt>SPECTRA-PKA</tt>, or can be used as a template to guide users who may want to create their own input data files (for example, for libraries not currently included on the download page). | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
== Example calculation == | == Example calculation == | ||
Latest revision as of 17:33, 23 November 2018
SPECTRA-PKA is a command-line driven programme for calculating the expected primary knock-on atom (PKA) spectra for a given target nuclide under neutron or charged particle irradiation. NJOY-processed recoil matrices must be provided as the input nuclear data for each nuclide and reaction channel of interest. SPECTRA-PKA will read the nuclear data and collapse the data for each reaction channel in a file with a user-defined energy spectrum of incident particles. SPECTRA-PKA outputs the resulting PKA spectra for each reaction channel read-in, as well as summed PKA distributions for each different recoiling nuclide or element, including both the typically heavy residuals and the secondary-emitted light gas particles. Crucially, SPECTRA-PKA has the ability to process data, in a single run, for a complex material containing many different target species. The user must specify the atomic percentage for each target and the appropriate recoil cross section file, but then SPECTRA-PKA will automatically read-in and process each file and create global average (including as a function of isotope and element) PKA distributions. This feature, in particular, is a significant advancement over what was possible before (such as with SPECTER [1985] from ANL), where typically PKA distributions were only provided for single elements (and not separated by reaction channel) and based on only one nuclear data library (ENDF/B-V in the case of SPECTER). It allows, for example, the user to investigate the variation in PKA distributions as a function of time under irradiation, where a materials composition may change due to transmutation.
Installation
SPECTRA-PKA comes with a simple makefile, requiring GNU make and a compatible Fortran compiler. It has been tested with multiple operating systems and compiler permutations, including the standard GNU Fortran compiler.
Usage
SPECTRA-PKA is executed from the command-line. By default, the programme expects to find an input file called specter.input in the execution folder, but this behaviour can be over-ridden by specifying the name of an input file on the command-line. The input file provides the user with the option to over-ride the default values of the various code words that SPECTRA-PKA relies upon to control its execution. The input file should be a text file but is largely free from other formatting requirements. Code word values are specified in any order, one per line, via syntax of type:
<code word name> = <code word value>
Code words
A complete list of code words (with explanation) can be found on the pdf readme on the git respository for SPECTRA-PKA. An alternative syntax allowing an easier description of a complex alloy (containing multiple target species) is also described.
Input PKA data download
SPECTRA-PKA uses NJOY-generated recoil cross section matrices. The output files required from NJOY are non-standard and are produced by a slightly modified version of the GROUPR module within NJOY. An evolving selection of databases produced using various international reaction cross section libraries (for various incident particle types) have been pre-calculated and are available to download as compressed tar archives from here (links are also provided in a detailed listing of nuclear data available for FISPACT-II at nuclear data downloads). These can be used immediately with SPECTRA-PKA, or can be used as a template to guide users who may want to create their own input data files (for example, for libraries not currently included on the download page).
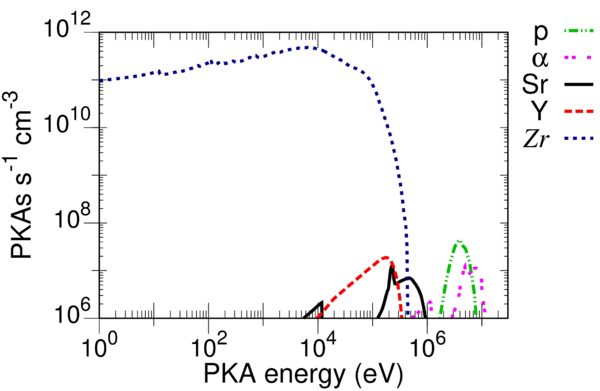
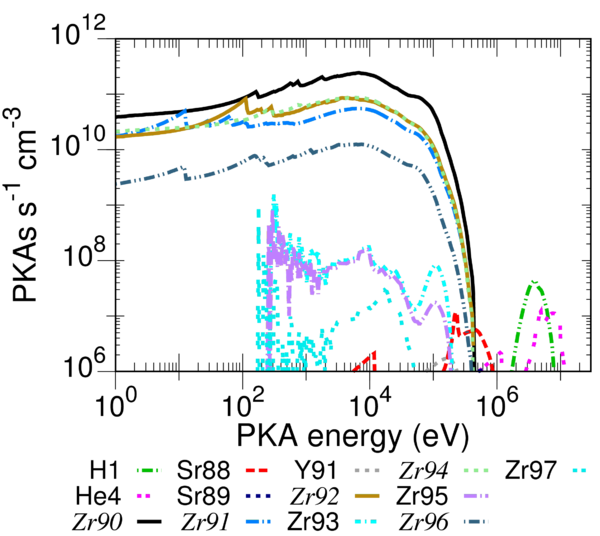
Example calculation
In the folder containing the distributed executables of SPECTRA-PKA there is a "test" folder to get the user started. The folder contains the necessary input files to evaluate the PKA distributions of pure zirconium (Zr) in a typical PWR fission spectrum. To run the test navigate to the test folder and then, on the command line, type:
flux_filename="fluxes_specter.dat" results_stub="ZR" num_columns=6 columns= pka_filename pka_ratios parent_ele parent_num ngamma_parent_mass ngamma_daughter_mass "<PKA data folder>Zr090s.asc" 0.51450000 Zr 90 89.904697659 90.905639587 "<PKA data folder>Zr091s.asc" 0.11220000 Zr 91 90.905639587 91.905034675 "<PKA data folder>Zr092s.asc" 0.17150000 Zr 92 91.905034675 92.906469947 "<PKA data folder>Zr094s.asc" 0.17380000 Zr 94 93.906310828 94.908038530 "<PKA data folder>Zr096s.asc" 0.02800000 Zr 96 95.908271433 96.910951206 flux_norm_type=2 pka_filetype=2 do_mtd_sums=.true. do_ngamma_estimate=.t. do_global_sums=.t. do_exclude_light_from_total=.t. number_pka_files=5 flux_rescale_value=3.25e14 max_global_recoils=400 energies_once_perfile=.t.
Note that in the test folder <PKA data folder> in the above has been replaced by the appropriate directory path for the default hierarchy of the FISPACT-II system. The user should tailor this file as necessary to their own system configuration before running the test.
The test folder also has an example_results folder containing the .out and .indexes files that should be produced from a successful execution of the test. The plot_example folder provides example .plt files for GNUPLOT that will produce plots of the summed elemental and isotopic PKA distributions. Users familiar with GNUPLOT will note the use of the index option in each plot command. The index numbers currently used in the .plt files are appropriate for the example results (see example_results/ZR.indexes). Important: Using a different version of the PKA source libraries could result in a different ordering of the results, and so the user should compare the .plt files with the .indexes file from their execution of the test to check that the correct distributions are plotted. The images that should result from this test are given in the figures shown.